

In zellulären Systemen stellt die DNA im Zellkern den zentralen Datenspeicher für vielfältige Funktionen dar. Neben der reinen Sequenz von DNA-Molekülen spielt jedoch die dreidimensionale räumliche Organisation der Mikro- und Nanoarchitektur eine wichtig Rolle zur Steuerung des komplexen Systems Zelle. Darüber hinaus findet man in den DNA-Sequenzdaten charakteristische Muster, die nicht zufällige Ordnungsprinzipien erwarten lassen und die in der Evolution entlang phylogentischer Linien hochgradig konserviert erscheinen.
Zelluläre Daten, die im Zellkern gespeichert sind, führen zu komplexen Funktionskaskaden und Regulationszyklen auf der Ebene der zellulären Proteine. Diese Funktionszyklen spiegeln sich häufig in Signalmolekülen auf der Zellmembran wider, so dass zelluläre Systeme mit ihrer Umgebung sich austauschen können. Gene und Nukleotidsequenzen im Zellkern und Proteine und Rezeptoren auf der Zellmembran können somit als korrespondierende Punkte von komplexen Funktionskaskaden eines Zellsystems gesehen werden. Molekular chemisch oder physikalisch getriggerte Veränderungen auf der einen wie auf der anderen Seiten führen zur räumlichen Umordnungen und Umverpackungen der DNA bzw. der Rezeptoren und Proteine, deren Dynamik und Charakteristika mittels moderner Methoden der hochauflösenden Mikroskopie und Nanomarkierung erforscht werden können.
Die Arbeitsgruppe „Experimentelle Biophysik“ beschäftigt sich unter Berücksichtigung biomedizischer und medizinisch diagnostischer Fragestellungen und Anwendungen mittels Verwendung von Fluoreszenzmikroskopie und hochauflösender Lokalisationsmikroskopie mit Fragen der Organisation von Biomolekülen und Zellbestandteilen in zellulären Systemen. Die Arbeiten dienen neben der Grundlagenforschung auch der angewandten Forschung in den Gebieten der Strahlenbiophysik und Tumormedizin.
Zur Erforschung des Informationsgehalts des Zellkernchromatins werden DNA Datenbanken auf spezische Muster analysiert und verglichen. Aus diesen Daten werden einerseits grundlegende Erkenntnisse der Chromatinorganisation gewonnen, andereseits können aus diesen Datenbanken auch Sequenzen von spezifischen Nanomarkern für die Mikroskopie auf der Basis von Oligonukleotiden bestimmt werden.
Solche und andere molekulare Markierungsmethoden erlauben es, sehr genaue Untersuchungen zu Strukturen und Organisationsprinzipien in dreidimensional erhaltenen Zellen durchzuführen. Dazu werden Zellen nicht nur äußeren Stimuli, wie ionisierender Strahlung, molekularen Liganden oder therapeutischen Antikörpern, sondern auch insbesondere geometrisch-physikalischen Randbedingungen ausgesetzt, die es ermöglichen, gewebenahe Zellensembles zu imitieren.
Die Erforschung von intrinsischer Information, die in Zellen archiviert und prozessiert wird, führt in einem interdisziplinären Diskurs zwischen Natur- und Geisteswissenschaften zu prinzipiellen Untersuchungen zur Organisation und Nutzung von Archiven im erweiterten Sinn.
Darüber hinaus werden aus theoretischen Betrachungen heraus Bedingungen bestimmt und untersucht, die für eine Entwicklung von lebensrelevanten molekularen Vorausetzungen auch unter extremen Randbedingungen möglich sind.
In der Arbeitsgruppe werden somit auf unterschiedlichen Ebenen der Komplexität und unter unterschiedlichen Randbedingungen zelluläre und subzelluläre Organisationstrukturen sowohl aus theoretischer wie auch aus experimenteller Sicht erforscht und in ein kohärentes biophysikalisches Bild zellulärer und subzelluläre Systeme zusammengefügt. Im einzelnen lassen sich die Arbeiten in folgende Projekte unterteilen:
Strahlenbiophysik: Spezische Inkorporation von Nanopartikeln (Nanogold) in Zellen und Zellkerne und Mechanismen der Wechselwirkung nach Strahlenexposition
Entwicklung von Konzepten zur nachhaltigen Archivierung biophysikalischer Daten
Astrobiophysik: Physikalische Randbedingungen zur Entwicklung lebender Systeme
Bachelor- / Masterarbeiten: Zu den laufenden Forschungsprojekten vergeben wir ständig Bachelor- und Masterarbeiten. Die Themen richten sich dabei nach aktuellen Fragestellungen, wobei auch persönliche Interessen und Neigungen berücksichtigt werden können. Falls Sie Interesse an unserer Arbeit haben, wenden Sie sich bitte an Herrn Prof. Hausmann oder einen seiner Mitarbeiter.
Die Arbeiten werden gefördert von:
Diese Seite wird gegenwärtig überarbeitet!
Etablierte Verfahren der FISH (Fluoreszenz In Situ Hybridisierung) nutzen als Sonden lange DNA-Sequenzen (> einige tausend Basen), die enzymatisch geschnitten und molekularbiologisch vermehrt werden. Die Lage der Markierungsregionen im Genom ist daher stark von der Lage der Schnittstellen bestimmt und führt insbesondere bei kleinen Genen zu weit über das Gen hinausgehende Markierungssonden.
Hinzu kommt, dass neue fluoreszenzmikroskopische Techniken, wie Lokalisationsmikroskopie, SNOM oder Wellenfeldmikroskopie etc.) in der Lage sind Strukturen des Chromatin auf der Nanometerskala zu vermessen, d.h. es werden einzelne Biomoleküle lokalisiert und deren Abstände bestimmt. Hierzu sind prinzipiell fluoreszente Sonden notwendig. Allerdings muss man bei Experimenten auf der Nanoskala berücksichtigen, dass diese Sonden aufgrund ihrer Größe keinen bzw. einen möglichst vernachlässigbaren Einfluss auf die native Struktur haben.
Kurze Oligonukleotide (< 50 Basen) lassen sich synthetisch in beliebiger Basenfolge sehr rein herstellen; allerdings ist bereits aus statistischen Gründen die Spezifität solcher Einzelsonden beschränkt. Die Kombination mehrerer solcher Sonden sollte jedoch eine spezifische Kolokalisation ermöglichen, so dass eine grundlegend andere Vorgehensweise erreicht werden kann, bei der der Aufbau von größeren Markierungsbereichen durch mehrere kleine DNA-Oligosequenzen erreicht wird. Damit ist das Problem der fokussierten Genmarkierung auf eine geeignete Kombinatorik von kleinen Oligonukleotiden reduziert. Gleichzeitig hat man Sonden, die sehr klein sind und entlang des Chromatinstrangs "Landmarken" setzen, aus denen eine Struktur berechnet werden kann.
An ausgewählten Beispielen aus der Tumordiagnostik (Bestimmung von Vielfachen von Genen), der Erforschung genetisch bedingter Erkrankungen (z.B. Fragiles X Syndrom) oder der Strahlenbiophysik (Untersuchung von Bruchereignissen der DNA) werden Kombinationen von Oligonukleotidsonden (typischerweise 15mere bis 30mere) zusammengestellt, die eine hochspezifische Fluoreszenzmarkierung der gewünschten Genloci mittels COMBO-FISH (COMBinatorial Oligonucleotide Fluorescence In Situ Hybridization) erlauben. Die Oligonukleotidkombinationen werden nach verschiedenen biophysikalischen Parametern so aus einer Genomdatenbank bestimmt werden, dass die Markierung auf den Genlokus im Zellkern fokussiert erfolgt und benachbarte Genombereiche nicht überdeckt werden. Damit kann gegenüber bisher etablierten Markierungsverfahren eine wesentlich höhere Spezifität und Selektivität der Markierung erreicht werden, so dass von Genkopienzahlveränderungen bis zu Strukturmessungen von Nanoschleifen genaue Untersuchungen möglich werden.
Durch die auf den Genlokus fokussierte Markierung aus spezifisch kolokalisierenden, kleinen Oligosonden ist es ferner möglich, die räumliche Ausdehnung eines einzelnen Genlokus im Zellkern durch hochauflösende Verfahren der Lichtnanoskopie genauer zu erfassen und in Relation zum Expressionsmuster der Zellen zu setzen. Schleifen und Netzwerke geeigneter Sondenmuster geben Auskunft über die nanoskalige Architektur des Zellkerns.
Weitere Informationen
Hausmann M, Cremer C. Verfahren zur mikroskopischen Ortsbestimmung eines ausgewählten, intrazellulären DNA-Abschnitts bekannter Nukleotidsequenz, Europäisches Patent, (EP 06006213.0), Anmeldetag 25. März 2006, erteilt am 13. 10. 2009.
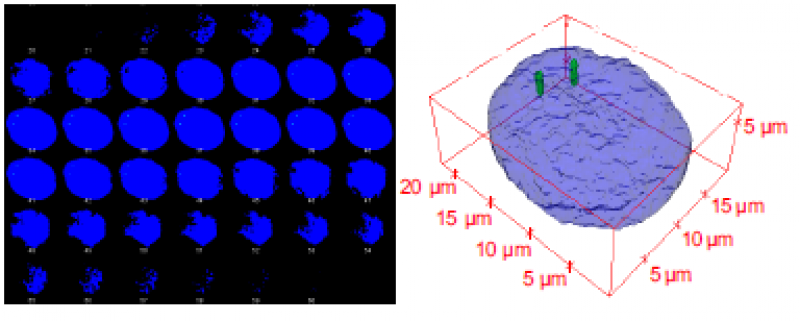
[ Serie von Bildschnitten nach konfokaler Mikroskopie und 3D Rekonstruktion eines Zellkerns. Das mit der Entstehung von Brustkrebs in Zusammenhang stehende Gen Her2/neu ist mittels 20 Oligonukleotid-Sonden spezifisch markiert. ]
In dem vom BMU geförderten Projekt der Strahlenbiophysik „3D/4D Architektur von chromosomalen Bruchpunktregionen in Zellkernen nach Bestrahlung von Krebs- und nicht-Krebszellen“, das gemeinsam mit Prof. Christoph Cremer (bis 31.12.2011: Forschungsgruppe „Angewandte Optik und Informationsverarbeitung“ am KIP, jetzt: Institut für Molekulare Biologie, Mainz) durchgeführt wird, geht es um die Erforschung der 3D Mikro- und Nanoarchitektur des Zellkerns und ihren Veränderungen nach Exposition ionisierender Strahlung.
Viele Forschungsergebnisse haben hierzu in den letzten Jahren gezeigt, dass das Genom im Zellkern sowohl auf der Mikro- als auch auf der Nanoskala nicht zufällig, sondern funktionell korreliert, organisiert ist. Nach Strahlenexposition unterliegt die 3D Architektur einer dynamischen Veränderung, die eng mit den jeweiligen Reparaturprozessen in Wechselwirkung steht. Ziel des Projektes ist es mit Hilfe neuer experimenteller Methoden und Computersimulationen (Kooperation D.W. Heermann, Institut für Theoretische Physik, Heidelberg) die Mechanismen zu erforschen, die die 3D Mikro- und Nanoarchitektur und ihre strahleninduzierten Veränderungen begründen. Die Kombination von spezifischen Mehrfarben-Fluoreszenz-Markierungsmethoden und hochauflösenden 3D Mikroskopie- und Lokalisationsnanoskopie-Techniken (SPDM = „Spatial Position Determination Microscopy“), die über lange Jahre am KIP entwickelt wurden, erlauben während des Ablaufs von Reparaturprozessen einen detaillierten Einblick in die supramolekulare und molekulare Organisation des Chromatins in Zellkernen zu erlangen. Auf der Ebene chromosomaler Cluster, wie z.B. Heterochromatin oder Euchromatin, als auch auf der Ebene ganzer Chromosomen werden die Auswirkungen von Strahlenexposition untersucht.
Das Projekt ist Teil eines Verbundes mit der Universität Darmstadt (Prof. M. Löbrich), der Universität der Bundeswehr, München (Prof. G. Dollinger), dem Klinikum der Ludwig Maximilians Universität, München (PD Dr. A. Friedl) und dem Helmholtzzentrum München (Dr. W. Friedland). Weitere Kooperationspartner sind: Institut für Radiobiologie der Bundeswehr assoziiert an der Universität Ulm (Prof. H. Scherthan) und dem Biomedical Research and Study Center, Riga, Lettland (Dr. J. Erenpreisa).
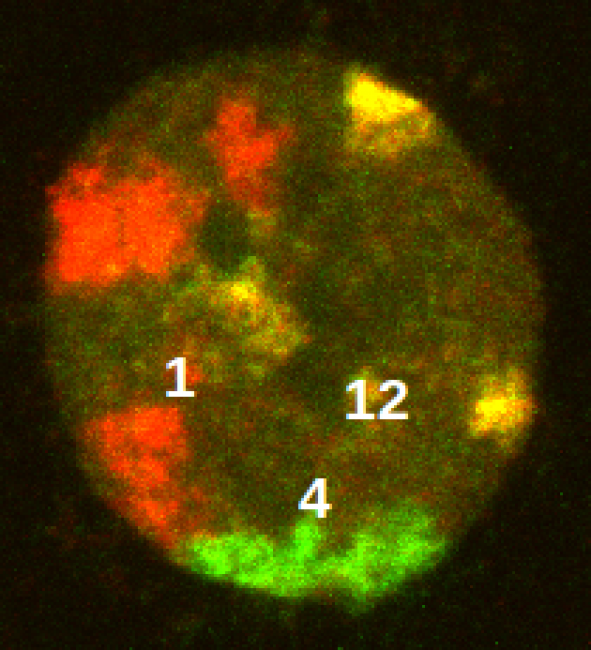
[ Beispiel eines Mikroskopbildes eines Zellkerns, in dem verschiedene Chromosomen spezifisch markiert sind. Bei Hochdosisbestrahlung versuchen die Zellen der mitotischen Katastrophe zu entgehen, indem sie ihre endopolyploide Genome umordnen (statistische Darstellung eines Beipiels) und diploide Zellen segregieren. ]
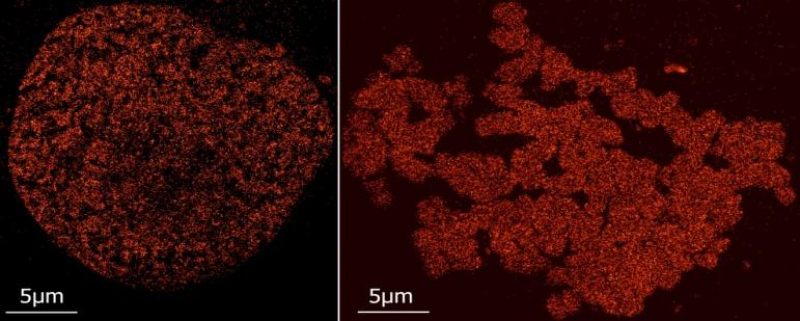
[ Darstellung eines Schnittbildes eines Zellkerns, bei dem die Nukleosomen spezifisch markiert wurden. Mittels SPDM-Lokalisationsmikroskopie wurden in einer Bildserie von mehreren hundert Bildern die Orte einzelner Moleküle, die durch Laserstrahlung zum Blinken angeregt werden können, bestimmt und im vorliegenden Bild rekonstruiert. Aus der Analyse der Punktmuster können Strukturen und ihre Veränderungen nach Strahlenexposition bestimmt werden. ]
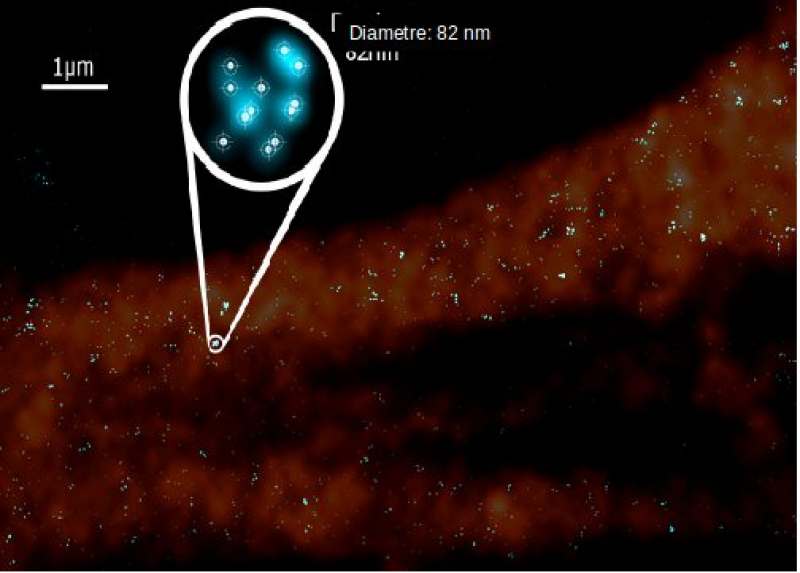
[ Darstellung eines Ausschnittes der Zellmembran, bei der erb-B2 mittels Antikörper markiert wurde. Die kreisförmige Ausschnittsvergrößerung zeigt, dass sich einzelne Punkte aus mehreren fluoreszenten Molekülen zusammensetzen, die Cluster bilden. Durch Präzisionsmessung der Abstände in den Clustern kann man die Zellen hinsichtlich der Tumorgenese klassifizieren und dynamische Veränderungen durch Stimulation von Expressionsmustern detektieren. ]
.jpg)
1. Solov’yov AV, Verkhovtsev AV, Mason NJ, Amos RA, Bald I, Baldacchino G, Dromey B, Falk M, Fedor J, Gerhards L, Hausmann M, Hildenbrand G, Hrabovský M, Kadlec S, Kočišek J, Lépine F, Ming S, Nisbet A, Ricketts K, Sala L, Schlathölter T, Wheatley AEH, Solov’yov IA (2024) Condensed Matter Systems Exposed to Radiation: Multiscale Theory, Simulations, and Experiment. Chem. Rev.: https://doi.org/10.1021/acs.chemrev.3c00902
2023